Our Research
We are a computational chemistry group comprised of myself and an ever changing group of undergraduate researchers . Currently most of our research is focused on fluorescent proteins, but we also model luciferases and methyl-coenzyme-M reductase.
Some Recent Papers:
Function and structure of GFP-like proteins in the protein data bank
Ong, W. J., Alvarez, S., Leroux, I. E., Shahid, R. S., Samma, A. A., Peshkepija, P., Morgan, A. L., Mulcahy, S., & Zimmer, M. (2011). Function and structure of GFP-like proteins in the protein data bank. Molecular bioSystems, 7(4), 984–992.
ABSTRACT: The RCSB protein databank contains 266 crystal structures of green fluorescent proteins (GFP) and GFP-like proteins. This is the first systematic analysis of all the GFP-like structures in the pdb. We have used the pdb to examine the function of fluorescent proteins (FP) in nature, aspects of excited state proton transfer (ESPT) in FPs, deformation from planarity of the chromophore and chromophore maturation. The conclusions reached in this review are that (1) The lid residues are highly conserved, particularly those on the “top” of the β-barrel. They are important to the function of GFP-like proteins, perhaps in protecting the chromophore or in β-barrel formation. (2) The primary/ancestral function of GFP-like proteins may well be to aid in light induced electron transfer. (3) The structural prerequisites for light activated proton pumps exist in many structures and it’s possible that like bioluminescence, proton pumps are secondary functions of GFP-like proteins. (4) In most GFP-like proteins the protein matrix exerts a significant strain on planar chromophores forcing most GFP-like proteins to adopt non-planar chromophores. These chromophoric deviations from planarity play an important role in determining the fluorescence quantum yield. (5) The chemospatial characteristics of the chromophore cavity determine the isomerization state of the chromophore. The cavities of highlighter proteins that can undergo cis/transisomerization have chemospatial properties that are common to both cis and trans GFP-like proteins.
Nwafor, J., Salguero, C., Welcome, F., Durmus, S., Glasser, R. N., Zimmer, M., & Schneider, T. L. (2021). Why Are Gly31, Gly33, and Gly35 Highly Conserved in All Fluorescent Proteins?. Biochemistry, 60(49), 3762–3770
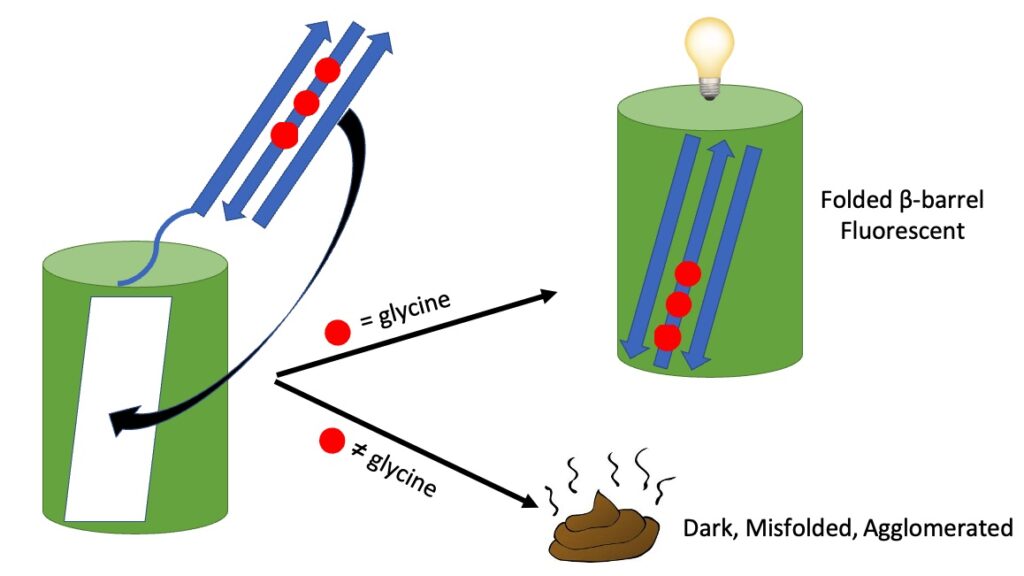
Abstract: Green fluorescent protein (GFP)-like fluorescent proteins have been found in more than 120 species. Although the proteins have little sequence identity, Gly31, 33, and 35 are 87, 100, and 95% conserved across all species, respectively. All GFP-like proteins have a β-barrel structure composed of 11 β-sheets, and the 3 conserved glycines are located in the second β-sheet. Molecular dynamics (MD) simulations have shown that mutating one or more of the glycines to alanines most likely does not reduce chromophore formation in correctly folded immature fluorescent proteins. MD and protein characterization of alanine mutants indicate that mutation of the conserved glycines leads to misfolding. Gly31, 33, and 35 are essential to maintain the integrity of the β1-3 triad that is the last structural element to slot in place in the formation of the canonical fluorescent protein β-barrel. Glycines located in β-sheets may have a similar role in the formation of other non-GFP β-barrels. Green fluorescent protein (GFP)-like fluorescent proteins have been found in more than 120 species. Although the proteins have little sequence identity, Gly31, 33, and 35 are 87, 100, and 95% conserved across all species, respectively. All GFP-like proteins have a β-barrel structure composed of 11 β-sheets, and the 3 conserved glycines are located in the second β-sheet. Molecular dynamics (MD) simulations have shown that mutating one or more of the glycines to alanines most likely does not reduce chromophore formation in correctly folded immature fluorescent proteins. MD and protein characterization of alanine mutants indicate that mutation of the conserved glycines leads to misfolding. Gly31, 33, and 35 are essential to maintain the integrity of the β1-3 triad that is the last structural element to slot in place in the formation of the canonical fluorescent protein β-barrel. Glycines located in β-sheets may have a similar role in the formation of other non-GFP β-barrels.
AlphaFold2 and RoseTTAFold predict posttranslational modifications. Chromophore formation in GFP-like proteins
Hartley, S. M., Tiernan, K. A., Ahmetaj, G., Cretu, A., Zhuang, Y., & Zimmer, M. (2022). AlphaFold2 and RoseTTAFold predict posttranslational modifications. Chromophore formation in GFP-like proteins. PloS one, 17(6), e0267560.
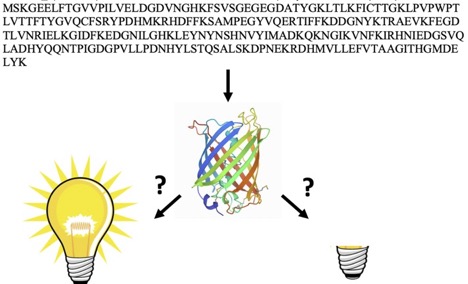
ABSTRACT: AlphaFold2 and RoseTTAfold are able to predict, based solely on their sequence whether GFP-like proteins will post-translationally form a chromophore (the part of the protein responsible for fluorescence) or not. Their training has not only taught them protein structure and folding, but also chemistry. The structures of 21 sequences of GFP-like fluorescent proteins that will post-translationally form a chromophore and of 23 GFP-like non-fluorescent proteins that do not have the residues required to form a chromophore were determined by AlphaFold2 and RoseTTAfold. The resultant structures were mined for a series of geometric measurements that are crucial to chromophore formation. Statistical analysis of these measurements showed that both programs conclusively distinguished between chromophore forming and non-chromophore forming proteins. A clear distinction between sequences capable of forming a chromophore and those that do not have the residues required for chromophore formation can be obtained by examining a single measurement-the RMSD of the overlap of the central alpha helices of the crystal structure of S65T GFP and the AlphaFold2 determined structure. Only 10 of the 578 GFP-like proteins in the pdb have no chromophore, yet when AlphaFold2 and RoseTTAFold are presented with the sequences of 44 GFP-like proteins that are not in the pdb they fold the proteins in such a way that one can unequivocally distinguish between those that can and cannot form a chromophore.
"The Role of the Tight-Turn, Broken Hydrogen Bonding, Glu222 and Arg96 in the Post-translational Green Fluorescent Protein Chromophore Formation.
N. P. Lemay*, A.L. Morgan*, E.J. Archer*, L.A. Dickson*, C.M. Megley* M. Zimmer Chemical Physics 2008, 348, 152-160.
ABSTRACT: Green Fluorescent Proteins (GFP) and GFP-like proteins all undergo an autocatalytic post-translational modification to form a centrally located chromophore. Structural analyses of all the GFP and GFP-like proteins in the protein databank were undertaken to determine the role of the tight-turn, broken hydrogen bonding, Gly67, Glu222 and Arg96 in the biosynthesis of the imidazolone group from 65SYG67. The analysis was supplemented by computational generation of the conformation adopted by uncyclized wild-type GFP. The data analysis suggests that Arg96 interacts with the Tyr66 carbonyl, stabilizing the reduced enolate intermediate that is required for cyclization; the carboxylate of Glu 222 acts as a base facilitating, through a network of two waters, the abstraction of a hydrogen from the alpha-carbon of Tyr66; a tight-turn conformation is required for autocatalytic cyclization. This conformation is responsible for a partial reduction in the hydrogen bonding network around the chromophore-forming region of the immature protein.
Photophysics And Dihedral Freedom Of The Chromophore In Yellow, Blue And Green Fluorescent Protein."
Colleen M. Megley, Luisa A. Dickson, Scott L. Maddalo, Gabriel J. Chandlera Marc Zimmer*Journal of Physical Chemistry B 2009, 113, 302-308.
ABSTRACT: Green fluorescent protein (GFP) and GFP-like fluorescent proteins owe their photophysical properties to an autocatalytically formed intrinsic chromophore. According to quantum mechanical calculations, the excited state of chromophore model systems has significant dihedral freedom, which may lead to fluorescence quenching intersystem crossing. Molecular dynamics simulations with freely rotating chromophoric dihedrals were performed on green, yellow and blue fluorescent proteins in order to model the dihedral freedom available to the chromophore in the excited state. Most current theories suggest that a restriction in the rotational freedom of the fluorescent protein chromophore will lead to an increase in fluorescence brightness and/or quantum yield. According to our calculations, the dihedral freedom of the systems studied (BFP > A5 > YFP > GFP) increases in the inverse order to the quantum yield. In all simulations the chromophore undergoes a negatively correlated hula-twist (also known as a bottom hula twist mechanism).
"Synergistic Mutations Produce Blue-Shifted Bioluminescence in Firefly Luciferase"
B. Branchini, D. Ablamsky, J. Rosenman*, L. Uzasci*, T. Southworth, M. Zimmer Biochemistry 2007, 46, 13847-13855.
ABSTRACT Light emission from the North American firefly Photinus pyralis, which emits yellow-green (557 nm) light, is widely believed to be the most efficient bioluminescence system known, making this luciferase an excellent tool for monitoring gene expression. In a previous study designed to produce luciferases for simultaneously monitoring two gene expression events, we identified a very promising blue-shifted emitter (548 nm) that contained the mutations Val241Ile, Gly246Ala, and Phe250Ser [Branchini, B. R., Southworth, T. L., Khattak, N. F., Michelini, E., and Roda, A. (2005) Red- and green-emitting firefly luciferase mutants for bioluminescent reporter applications, Anal. Biochem. 345, 140-148]. To establish the basis of the unusual blue-shifted emission, we determined that a simple additive effect of the three individual mutations did not account for the spectral properties of the triple mutant. Instead, the bioluminescence emission spectra of two double mutants containing Phe250Ser and either Val241Ile or Gly246Ala very closely resembled that of the triple mutant. Additional mutagenesis results confirmed that the blue-shifted emission of the double mutants was determined by the synergistic behavior of active site residues. Molecular modeling studies of the Gly246Ala and Phe250Ser double mutant supported the notion that the blue-shifted emission was due to localized changes that increased the hydrophobicity at the emitter site as a result of the addition of a single methyl group at position 246. Moreover, the modeling data suggested that the Ala246 side chain remained close to the emitter through an additional H-bond between Ala246 and the hydroxyl group of Phe250, providing a possible structural basis for the synergistic behavior.
"Are classical molecular mechanics calculations still useful in bioinorganic simulations?
M. Zimmer Coordination Chemistry Reviews in press.
ABSTRACT There are currently no widely distributed molecular mechanics programs with inorganic force fields that allow the user to accurately model the structure of bioinorganic molecules without a prior parameterization of the metal coordination sphere. However, there are still times when the speed, accuracy and output of the calculations make inorganic molecular mechanics (MM) the method of choice. Bioinorganic molecular mechanical calculations are most commonly used to examine metalloproteins or parts of metalloproteins that are too large for analysis by QM/MM or DFT; to search large areas of conformational space; or in molecular dynamics. To illustrate the utility of inorganic molecular mechanics in bioinorganic chemistry, a brief review of simulations of bioinorganic compounds containing transitional metals published in the last 5 years is presented together with a computational analyses of methyl coenzyme-M reductase (MCR), which describes calculations that are too large for QM based methods, and urease, which is used as an example to illustrate how MM calculations can be used to sample large areas of conformational space. In the case of MCR inorganic molecular mechanical conformational searches in conjunction with hole scans and normal-coordinate structural decomposition analysis are used to show how the protein matrix inhibits the non-planar deformations of the tetrapyrrole cofactor F430 and thereby changes its redox chemistry – an entatic effect. In our analysis of urease inorganic molecular mechanical conformational searches were used to examine the conformational space available to the substrate and urea, and to examine potential mechanisms for its degradation.
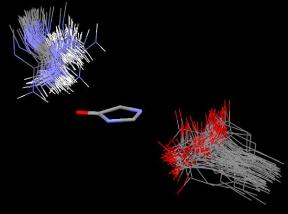
Figure 4. An Isostar overlay plot of all the crystal structures of GFP and GFP-like proteins with fully cyclized chromophores found in the pdb (99 structures, see supplementary Table I for list of structures). The imidazolone rings of all the structures were overlapped in order to show the orientation of the arginine and glutamic acid residues (Arg96 and Glu222 in GFP numbering) relative to the position of the imidazolone ring.
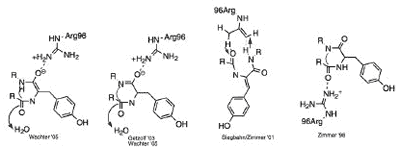
Figure 6. Possible roles for Arg96 in the autocatalytic cyclization reaction responsible for chromophore formation.