Spinach: An RNA mimic of GFP
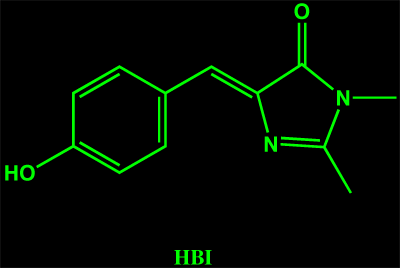
Numerous studies of green fluorescent protein’s photochemistry have utilized model compounds synthesized to mimic the proteins chromophore. The most commonly used is HBI, shown on the left.
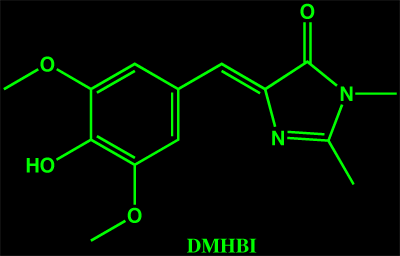
These organic model compounds only fluoresce when their dihedral freedom is severely restricted. Jaffrey and co-workers(1) have designed two new ligands based on HBI, DMHBI and DMFBI (both shown here).
These fluorophores selectively bind specific RNA sequences that were found by ten rounds of systematic evolution of ligands by exponential enrichment (SELEX). When bound to the RNA aptamer their dihedral freedom is sufficiently restricted that they fluoresce. Like GFP, DMHBI is predominantly found in the phenolic form, the brighter DMFBI mimics EGFP and is anionic under standard conditions. The DMFBI-RNA complex has been called Spinach, it is the first fluorescent RNA tag that is both selective and non-toxic. It only fluoresces when bound to RNA, is resistant to photobleaching and has been used to follow RNA tagged molecules as they move through cells.